Progress in the RERF Biochemical Genetics Study
The ongoing hunt for heritable mutations among the children of the atomic-bomb survivors will involve surveying large numbers of nucleotides for various types of mutation.
by Chiyoko Satoh and Mieko Kodaira, Department of Genetics, RERF
This is a revision of an article that originally appeared in RERF Update 6(3):3-4, 1994. The most recent data concerning this research have been published in: Satoh C, Takahashi N, Asakawa J, Kodaira M, Kuick R, Hanash SM, Neel JV. Genetic analysis of children of atomic bomb survivors. Environmental Health Perspectives 104(Suppl 3):511-9, 1996.
To study at the DNA/RNA level the possible genetic effects on the children of atomic-bomb (A-bomb) survivors, in the RERF Laboratory of Biochemical Genetics, we are developing technologies and establishing cell lines that will be used in future screening projects.
Establishing cell-line repositories
Our goal is to establish cell lines from B lymphocytes from 1000 families using Epstein-Barr virus transformation. Resorting to cell lines was deemed necessary because in mutation studies, both parents of a child must be available, and by now–almost 50 years after the bombings–the parents of many first-generation children are unavailable due to death or emigration. Among 500 of these families, one or two parents were exposed to A-bomb radiation, and the combined parental dose is more than 0.01 Sv; the average combined parental dose for these families is 0.5 Sv. The other 500 families are controls. Each family comprises a mother, a father, and all available children.
Each established cell line is proliferated to between 5 × 108 and 1 × 109 cells, from which an adequate amount of DNA can be extracted for various analyses, and each cell line is preserved in liquid nitrogen. Aliquots of intact lymphocytes and granulocytes are also preserved in liquid nitrogen as reference materials to determine if mutations encountered during screening occurred during cell-line establishment.
To date, cell lines have been established for more than 500 families in Hiroshima and for more than 440 families in Nagasaki, where the project started one year later. Because samples were selected on the basis of the revised tentative 1965 dosimetry, some parents do not have Dosimetry System 1986 (DS86) dose estimates. Thus, we are planning to add some new families that have parental DS86 doses.
Determining potential DNA targets
As reported in RERF Update 3(4):1, 1991, at a workshop on human germline mutagenesis held at the Hiroshima Laboratory in November 1991, workshop attendees recommended a pilot study to compare various types of DNA as potential targets for detecting germinal mutations.
In the middle of 1992, we began to examine 100 families–a subset of the 1000 families used in establishing our cell lines. Various genes or DNA sequences have been examined using three techniques: (1) denaturing gradient gel electrophoresis of the products of polymerase chain reaction (PCR) for detecting single nucleotide substitutions and small deletions and insertions in DNA or RNA fragments of 2-3 kilobase pairs (kb), (2) Southern blotting for detecting rather long sequence differences made from insertions/deletions/rearrangements (I/D/R) in unique sequences and for detecting changes in the number of repeats in minisatellites or variable number of tandem repeats (VNTRs), and (3) high-resolution electrophoresis of PCR-amplified microsatellites for detecting the difference in the numbers of unit sequences in microsatellites. In this article, we will report on our examination of minisatellites and microsatellites.
Exploring the mini- and microsatellites
Minisatellites or VNTRs are dispersed throughout the human genome but show a tendency to cluster in telomeric regions. They consist of short-sequence units (~5-40 base pair [bp] in length) iterated in tandem to form arrays of approximately 1-10 kb and show allelic variation in the number of repeat units. At several minisatellite loci that exhibit high heterozygosity, high germline mutation rates for new length alleles were observed. We examined six minisatellite loci from 50 exposed families including 64 children and 50 control families with 60 children. Among the 64 children from the exposed families, only one child has parents who were both exposed. Thus, by examining 124 children of the 100 families, we could examine 65 gametes (sperm and oocytes) produced in exposed parents and 183 gametes derived from unexposed parents (63 gametes from the unexposed parents in the exposed families plus 120 from the control parents). The average parental gonad dose for the 65 gametes was 1.9 Sv.
DNA samples extracted from the cell lines were digested by HinfI restriction enzyme, which does not cleave within the minisatellite tandem-repeat array, and conventional agarose gel electrophoresis was carried out. Southern filters produced from the agarose gels were sequentially hybridized with six minisatellite probes. We compared the children’s bands with their parents’ bands and identified mutant bands when bands of identical length were absent in both parents. When mutants were detected, we examined the DNA samples from these families after digestion by a second restriction enzyme, AluI, that cut outside of the repeat array at positions different from HinfI digestion sites. An example of a mutation detected by the CEB-1 probe is shown in the Figure. A decrease in mobility of the son’s band as compared to that of his father’s band by approximately 2 kb in the HinfI digest was reproduced in the AluI digest, which is consistent with the idea that the mutation involves the loss of numbers of repeat units within the minisatellite. To exclude the possibility that the mutations had been generated during cell-line establishment, we examined DNA from intact lymphocytes from the members of those families in whom mutations had been detected. In all mutation cases, we observed mutant bands identical to those observed in the cell-line DNA of the children, confirming that the mutations had occurred in the parents’ germ cells.
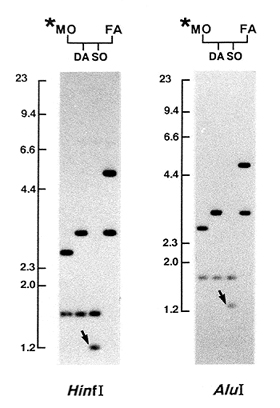
Figure. A new length mutation at the CEB-1 locus detected in a child of Family 0716. Arrows indicate mutant bands in the digests of HinfI and AluI, respectively. One of the son’s (SO) bands is identical to one of the mother’s (MO) bands, but the other band is identical to neither of the father’s (FA) bands. DA = daughter. The asterisk indicates that the mother was exposed to atomic-bomb radiation. Positions and lengths (in kilobase pairs) of DNA markers are at the left of the photographs.
We detected six mutations after examining 65 gametes at three minisatellite loci ( MS1,CEB-1, p
MS1,CEB-1, p g3); no mutations were detected at the remaining three loci. Interestingly, 22 mutations were detected at the same three minisatellite loci in the 183 unexposed gametes. The average mutation rates (mutation/loci/gamete) for the exposed and the unexposed gametes were 0.015 [6 ÷ (65 × 6)] and 0.02 [22 ÷ (183 × 6)], respectively. The average mutation rates at the six minisatellite loci of the gametes derived from the exposed parents, and the unexposed parents did not show a statistically significant difference. Moreover, at each of the three minisatellite loci where mutations had been detected, the mutation rate did not differ significantly between the exposed and unexposed gametes.
g3); no mutations were detected at the remaining three loci. Interestingly, 22 mutations were detected at the same three minisatellite loci in the 183 unexposed gametes. The average mutation rates (mutation/loci/gamete) for the exposed and the unexposed gametes were 0.015 [6 ÷ (65 × 6)] and 0.02 [22 ÷ (183 × 6)], respectively. The average mutation rates at the six minisatellite loci of the gametes derived from the exposed parents, and the unexposed parents did not show a statistically significant difference. Moreover, at each of the three minisatellite loci where mutations had been detected, the mutation rate did not differ significantly between the exposed and unexposed gametes.
We have estimated the number of germ cells needed to demonstrate a significant difference between the mutation rates of the exposed and the unexposed germ cells. For a locus with a spontaneous mutation rate of 0.02, identical to that of the mean mutation rate of the six loci examined in our study, it would require two samples of 1188 germ cells (Kodaira et al., Am. J. Hum. Genet. 57:1275-83, 1995). This means we have to examine 19 loci from each of these 64 children from the 50 exposed families, ie, 13 additional loci with a mutation rate of 0.02/gamete/loci are necessary. We think this is a practical number. We now are looking for new minisatellite loci with spontaneous mutation rates greater than 0.02 for additional screening. Multilocus-minisatellite probes that detect many hypervariable minisatellite loci simultaneously and that also are being used for DNA fingerprinting may be suitable probes for our purpose.
Microsatellites consist of around 10-50 copies of motifs from 2 bp to 6 bp. They are highly polymorphic in the copy number of motifs, are randomly distributed in human DNAs, and occur frequently. The aberrant expansion of exonic trinucleotide repeats has recently been found to result in genetic diseases, such as fragile X syndrome, myotonic dystrophy, spinobulbar muscular atrophy (SBMA), and Huntington’s disease, and represents a new form of mammalian mutagenesis. In the FMR-1 gene responsible for the fragile X syndrome, a CGG-repeat in the 5′-untranslated region is normally polymorphic, displaying alleles ranging from 6-54 repeats, but affected individuals have more than 200 copies. In addition, high spontaneous mutation rates (~0.013-0.015) at the tetranucleotide repeats in some of the genes in Caucasian families and instability of microsatellites in hereditary nonpolyposis colorectal cancer have been reported.
We examined the numbers of trinucleotide repeats in the FMR-1, AR (the gene responsible for SBMA), and DM genes of 124 children, the former two genes being on the X chromosome. Sequences including microsatellites were amplified by PCR, and products were electrophoresed on a sequence gel. Bands from these children were compared with those of their parents. A total of 177 alleles from the children with triplet repeats derived from the exposed parents, and 443 alleles derived from the unexposed parents were screened for mutations. No mutations were detected among 620 alleles in the children. We now are examining several tetranucleotide repeats and soon can determine whether these microsatellites are suitable targets.
Two-dimensional gel electrophoresis of DNA
The fourth technique that will be introduced into the pilot study is known as two-dimensional gel electrophoresis (2-DE). Using 2-DE, approximately 2000 restriction fragments of DNA from an individual can be detected on a gel. The beauty of this method is that no probes are necessary, and it can detect deletions and insertions as well as base-pair substitutions at the restriction sites. Because these events generally occur only in one of the two chromosomes–the remaining chromosome being normal–spots of DNA fragments from affected individuals will be detected at the normal position with an intensity 50% less than the intensity of spots from unaffected individuals. Spots of DNA fragments resulting from deletions, insertions, or base-pair substitutions will disappear, increase their intensities, or move to other positions. Thus, to detect these events occurring in heterozygotes for an affected allele and a normal allele, accurate quantitation–ie, the ability to detect a 50% decrease or increase in the intensity of the spots–is necessary. The gel quality has reached a level such that the electrophoresis patterns derived from a single DNA sample exhibit distribution patterns of spots that can be superimposed.
Two-dimensional gel electrophoresis of DNA samples from three mother/father/child trios has been done in our laboratory by research scientist Jun-ichi Asakawa, and autoradiograms were analyzed using a computer-assisted image analyzer at the University of Michigan Medical School, where software developed for SM Hanash and James V Neel has proved to be effective in quantitating the intensity of the spots.
We hope to report on 2-DE analysis in an upcoming issue of RERF Update (7[1]:3-5, 1995).

